How to Identify Which Genes Have Dose Response Using R
Genomic approaches to SADRs. Efficacy threshold C Gene clusters app.

Genetic Basis Of Variation In Cocaine And Methamphetamine Consumption In Outbred Populations Of Drosophila Melanogaster Pnas
Column 4 is the gene lengthnum_genes 2.

. In reality Several different routes Exposure to more than one chemical at a time. The gene can then be studied further by molecular biology techniques such as cloning and. Combined dependency score profile of the gene selected in 2.
Whole-plant dose-response experiments were carried out to determine the GR 50 values of the purified R AH18 and confirmed S SD01 populations to mesosulfuron-methyl in the absence and presence of malathion. However response to concentration may be complex and is often nonlinear. Low dose patients and the compliment.
This study identified a wide array of druggable genes for both lung and head and neck cancer. DNA sequences that code for proteins begin with the three bases ATG that code for the amino acid methionine and they end with one or more stop codons. We found that this assay was less time consuming more sensitive and higher throughput than gene expression analysis.
However high dose groups 50 mGyyear have higher representationabundance of genes involved in DNA damage response that include DNA repair response to oxidative stress immune response chromatin modification methylation and histone modification mRNA processing cell cycle and apoptosis Fig 6. Querying a sequence. Evidence for a familial or genetic.
The gene or genes that have been altered in the mutant organism are then studied by genetic crosses Section 524 which can locate the position of a gene in a genome and also determine if the gene is the same as one that has already been characterized. The level of evidence required to adopt a specific genetic test as a guide to practice is not established. I wantto use the t- test to identify the DEGs.
Viewed 874 times -2 I have a matrix which contains the genes and the mrna. Use of genotyping is more accurate than race or ethnic categories to identify variations in drug response52 Unlike other influences on drug response genetic factors remain constant throughout. The rnr gene that encodes RNase R is part of an operon containing.
Genes where no reads have mapped. The relationship between the drug dose regardless of route used and the drug concentration at the cellular level is. Bioinformatics allows scientists to make educated guesses about where genes are located simply by analyzing sequence data using a computer in silico.
Dropdown menu to select a gene symbol that matches the query. Modified 9 years 7 months ago. In particular multiple genes involved in the G2-M checkpoint were shown to be essential for tumor cell survival indicating their potential as anticancer targets.
The genetic make-up of the population is not very diverse. How would it be possible to find the differentially expressed genes in such a case. You will now most likely unless you have bacterial data.
Dose-dependent increases in intracellular fluorescent phospholipid accumulation were observed. We identify 8 novel associations after bonferroni correction 3 of which are replicated or validated in the UK biobank or have other supporting results. Chronic Toxicological Studies 2 Characteristics of animal studies A single dose A single route of exposure The number of animals exposed is small.
The plot allows us to identify the doses that affect a percentage of the exposed population. Dropdown menu to select an. But do not know how to proceed with four different types of promoters and identify the DEGs.
The distributions of gene-scores of the two classes were tested for the null hypothesis namely that the means of the distributions were equal. The genes data is a num_genes x 4 matrix where num_genes is the total number of uniquely annotated genes. Most data sets available contains a lot of 0-measurements see for instance your density plot from the previous chapter.
If Column 2 1 and Column 3 4 then exons 123 and 4 are inside the gene. ID_REF GSM362168 GSM362169 GSM362170 GSM362171 GSM362172 GSM362173 GSM362174 244901_at 5171072 5207896 5191145 5067809 5010239. I know to find the DEGs using the multtest package in R in case of two conditions such as control and treatment.
Using an expanded set of compounds we show that this assay correctly identified 100 of PLD-positive and -negative compounds. 14 Operon transcription is thought to be driven by a σ 70 promoter and transcript processing and decay is mainly regulated by RNase E. This combination therapy may have translational value for patients with TNBC and improve therapeutic response for this aggressive form of breast cancer.
The gene-score was calculated for each of the 228 genes in the WSP for each patient in two classes. Cross-species genomic and functional analyses identify a combination therapy using a CHK1 inhibitor and a ribonucleotide reductase inhibitor to treat triple-negative breast cancer. T test to find differentially expressed genes in R.
The C-allele at rs4918758 in CYP2C9 was associated with a 25 15-44 lower odds of dose reduction of quinine p1610-5. B Gene essentiality app. List of matched genes.
The A-allele at rs9895420 in ABCC3 was associated with a 46 24-62. Column 1 is the gene ID column 2 is the starting exon and column 3 is the ending exon in the gene. 189 RNase R levels change in response to different.
Further consensus recommendations on dose adjustment may not be available even when large effects of genetic variants on drug response have been identified. Protein and gene sequence comparisons are done with BLAST Basic Local Alignment Search Tool. Extreme dose response warfarin response models.
Textbox to type in a partial gene symbol to query. In principle locating genes should be easy. Identifying genetic risk factors for SADRs particularly Type B reactions could significantly decrease the healthcare costs and improve the process of drug development 5Characteristics of SADRs that increase the likelihood of informative genetic or genomic analysis include.
In the R-studio Environment tab click on your data set or perform the Viewdata command and click on one of the sample columns to order the data ascending. Occursthrough binding or chemical interactionthe concentration of the drug at the site of action controls the effect. Ask Question Asked 9 years 7 months ago.
A dose can be described either as a lethal dose LD in which the response is the death of animals or cells or an effective dose ED in which the response is another observable outcome. To access BLAST go to Resources Sequence Analysis BLAST. This is an unknown protein sequence that we are seeking to identify by comparing it to known protein sequences and so Protein BLAST should be selected from the.
The malathion used in this experiment was formulated in a mixture of a commercial solvent and an emulsifier. Only individuals in good health andor of the same sex are selected. NsrR transcriptional regulator rnr exoribonuclease rlmB rRNA methyltransferase and yjfI unknown function.

133 Questions With Answers In Graphpad Prism Science Topic

Integrated Genome And Transcriptome Analyses Reveal The Mechanism Of Genome Instability In Ataxia With Oculomotor Apraxia 2 Pnas
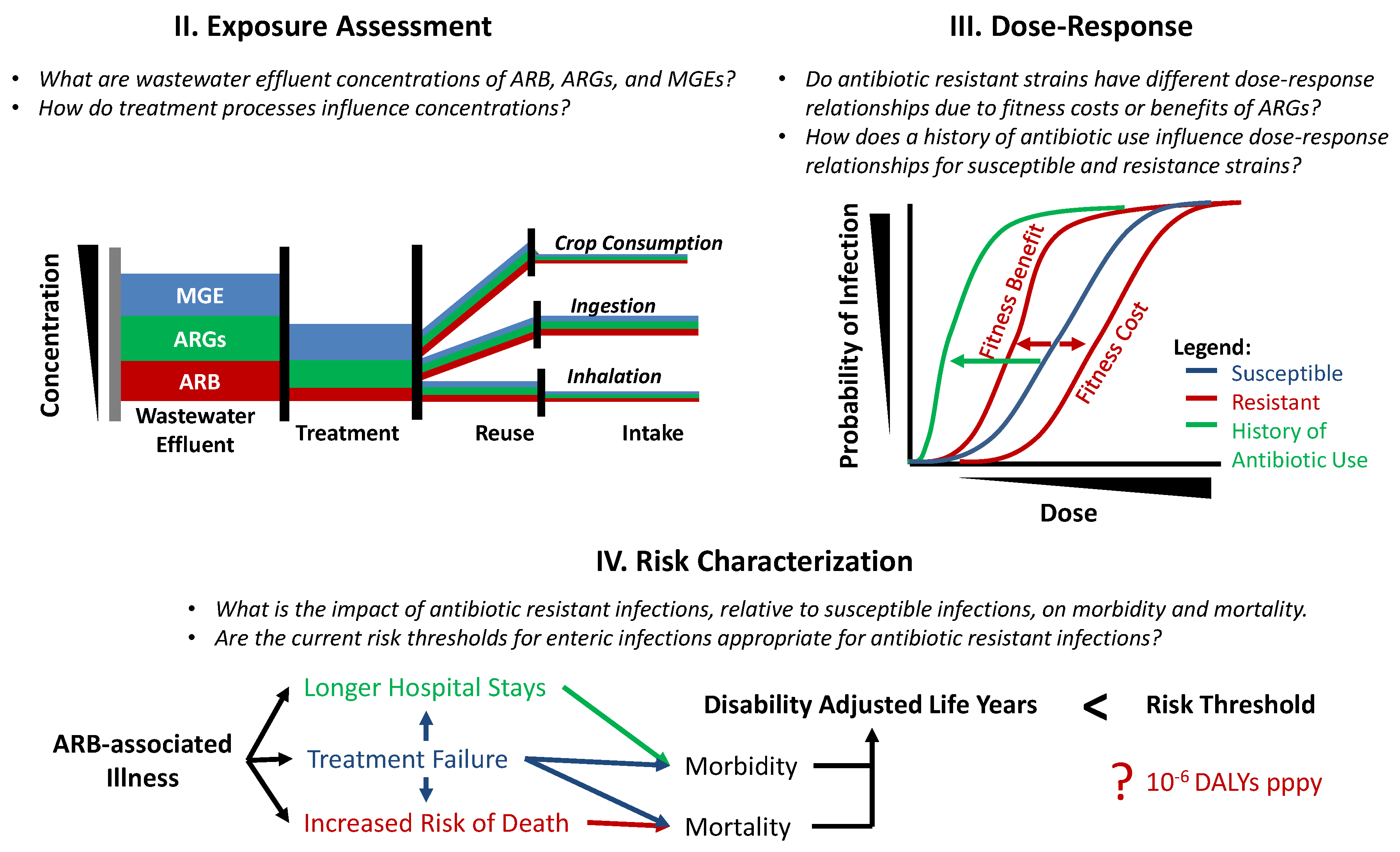
Water Free Full Text Reusing Treated Wastewater Consideration Of The Safety Aspects Associated With Antibiotic Resistant Bacteria And Antibiotic Resistance Genes Html
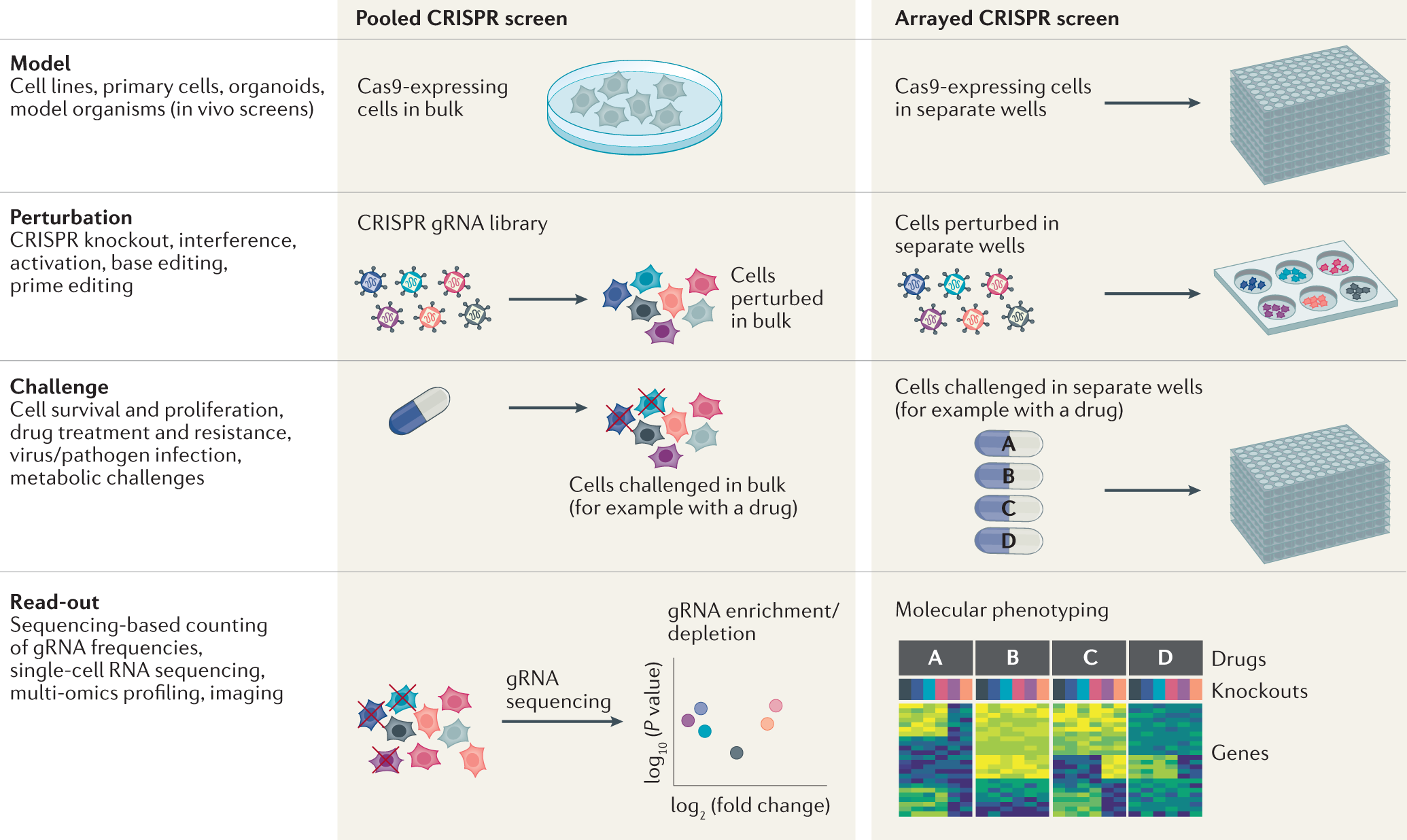
High Content Crispr Screening Nature Reviews Methods Primers

Exome Sequencing Identifies Rare Coding Variants In 10 Genes Which Confer Substantial Risk For Schizophrenia Medrxiv
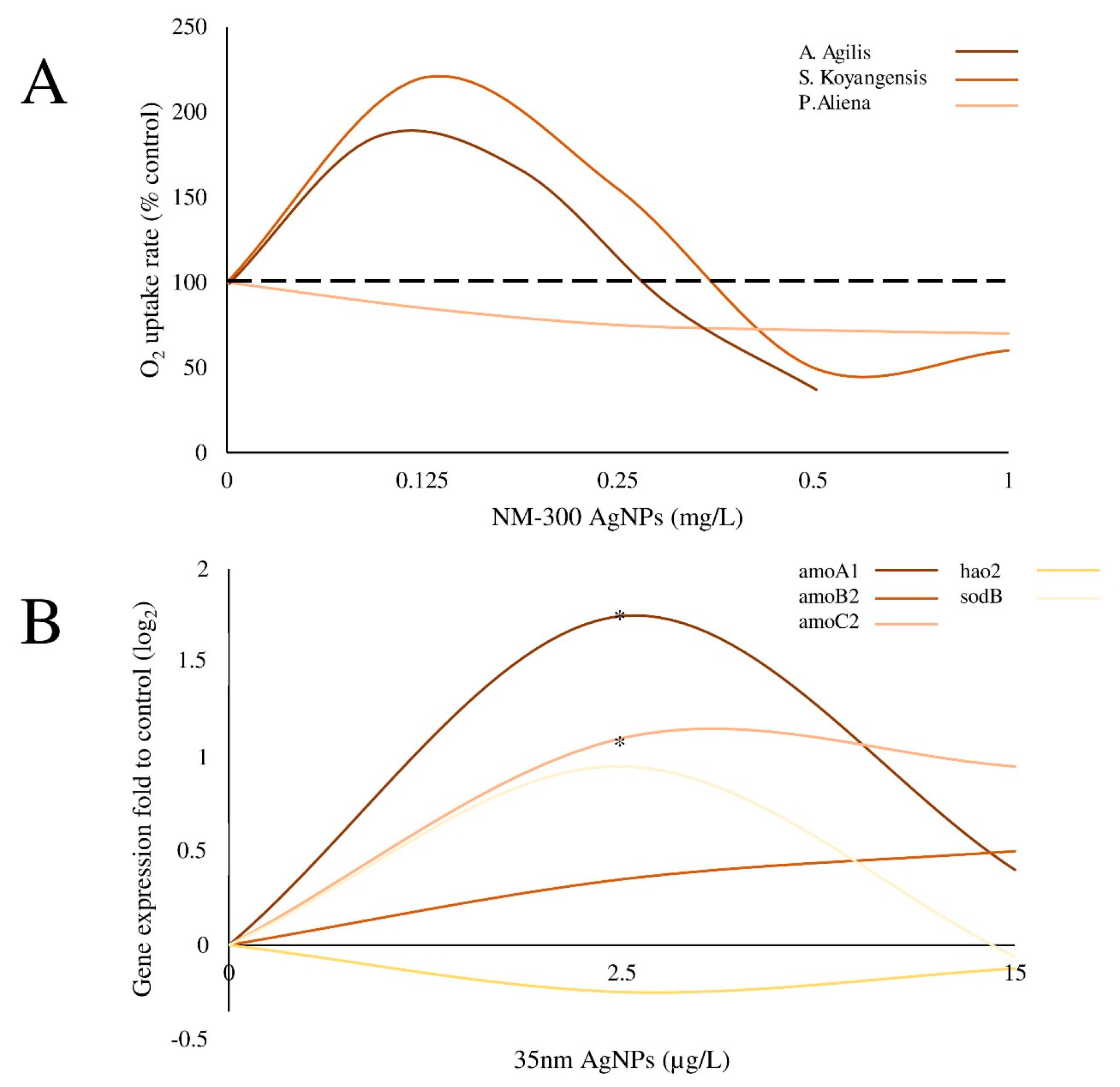
Ijms Free Full Text Nanoparticle Exposure And Hormetic Dose Responses An Update Html

A Pipeline To Analyse Time Course Gene Expression F1000research

A Chemical Genetic Screen Identifies Aurora Kinases As A Therapeutic Target In Egfr T790m Negative Gefitinib Resistant Head And Neck Squamous Cell Carcinoma Hnscc Ebiomedicine

Computational Methods For Identifying Similar Diseases Molecular Therapy Nucleic Acids

Identification Of Covid 19 Prognostic Markers And Therapeutic Targets Through Meta Analysis And Validation Of Omics Data From Nasopharyngeal Samples Ebiomedicine
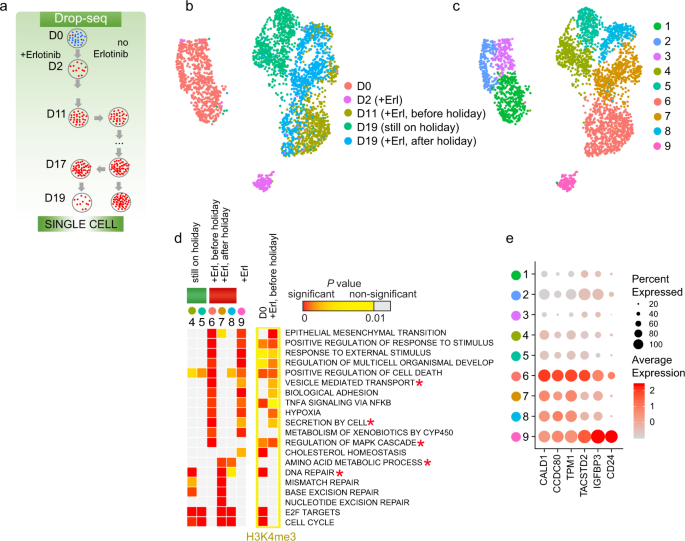
Single Cell Transcriptional Changes Associated With Drug Tolerance And Response To Combination Therapies In Cancer Nature Communications
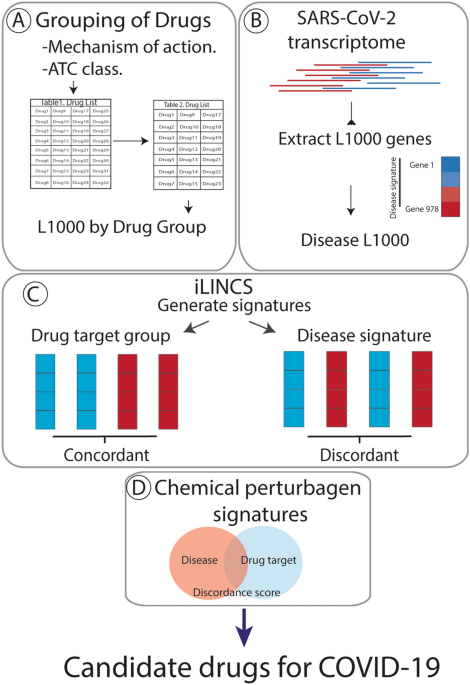
Identification Of Candidate Repurposable Drugs To Combat Covid 19 Using A Signature Based Approach Scientific Reports

A Pipeline To Analyse Time Course Gene Expression F1000research
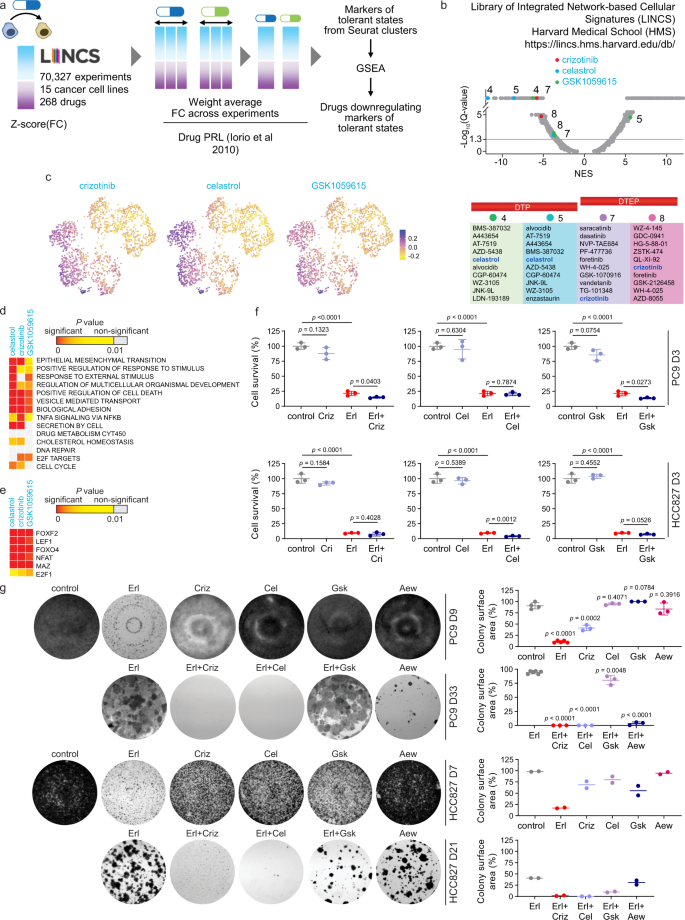
Single Cell Transcriptional Changes Associated With Drug Tolerance And Response To Combination Therapies In Cancer Nature Communications

Deep Learning Predicts Gene Expression As An Intermediate Data Modality To Identify Susceptibility Patterns In Mycobacterium Tuberculosis Infected Diversity Outbred Mice Ebiomedicine

Genome Wide Analysis Of Therapeutic Response Uncovers Molecular Pathways Governing Tamoxifen Resistance In Er Breast Cancer Ebiomedicine

Genome Wide Analysis Of Therapeutic Response Uncovers Molecular Pathways Governing Tamoxifen Resistance In Er Breast Cancer Ebiomedicine
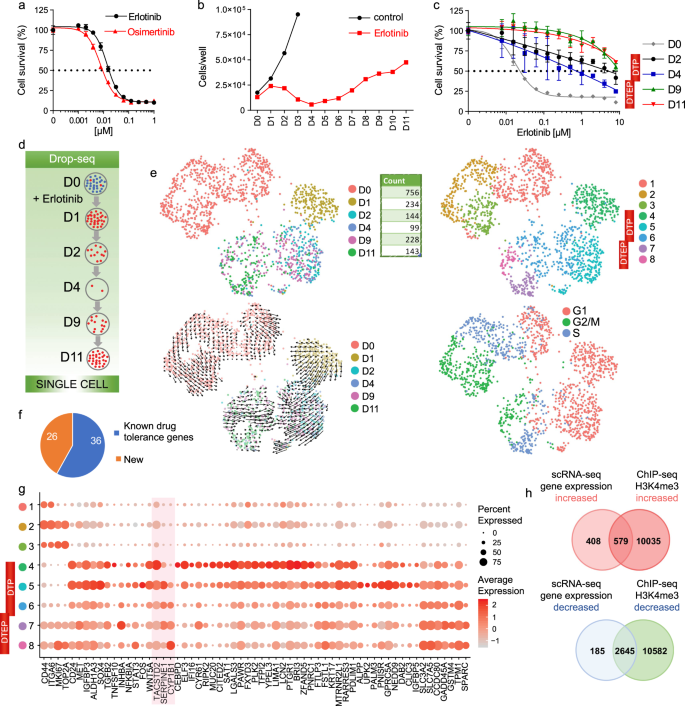
Single Cell Transcriptional Changes Associated With Drug Tolerance And Response To Combination Therapies In Cancer Nature Communications
Comments
Post a Comment